cclib + chemlab for orbital functions¶
Dev code, see - http://localhost:8890/lab/tree/MO_tests_cclib-chemlab_060420.ipynb - http://localhost:8889/notebooks/github/ePSproc/epsproc/tests/plottingDev/cclib_chemlab_ITK_pyVista_dev_150420.ipynb
Here: test packaged fns.
Imports¶
[1]:
!hostname
!conda env list
bemo
# conda environments:
#
base /home/femtolab/anaconda3
chem /home/femtolab/anaconda3/envs/chem
chem3.5 /home/femtolab/anaconda3/envs/chem3.5
dbtest /home/femtolab/anaconda3/envs/dbtest
epsdev * /home/femtolab/anaconda3/envs/epsdev
epsdev-150320 /home/femtolab/anaconda3/envs/epsdev-150320
espressoDB /home/femtolab/anaconda3/envs/espressoDB
fibre-sim /home/femtolab/anaconda3/envs/fibre-sim
remote /home/femtolab/anaconda3/envs/remote
slides /home/femtolab/anaconda3/envs/slides
webDev /home/femtolab/anaconda3/envs/webDev
[2]:
import sys
import os
import numpy as np
# For module testing, include path to module here
modPath = r'/home/femtolab/github/ePSproc'
sys.path.append(modPath)
import epsproc as ep
# import epsproc as ep
* plotly not found, plotly plots not available.
* pyevtk not found, VTK export not available.
Submodule testing¶
[3]:
from epsproc.vol import orbPlot
[4]:
dir(orbPlot)
[4]:
['Path',
'__builtins__',
'__cached__',
'__doc__',
'__file__',
'__loader__',
'__name__',
'__package__',
'__spec__',
'cclib',
'importChemLabQC',
'molOrbPlotter',
'np',
'pv',
'sys']
[5]:
orbPlot.importChemLabQC()
Chemlab orbital functionality not found, orbital calculations not possible.
[6]:
chemPath = r'/home/femtolab/python/chem/tools/chemlab/chemlab'
out = orbPlot.importChemLabQC(chemPath = chemPath)
Import OK: Chemlab module <module 'qc.wavefunction' from '/home/femtolab/python/chem/tools/chemlab/chemlab/qc/wavefunction.py'>
[7]:
dir(orbPlot)
[7]:
['Path',
'__builtins__',
'__cached__',
'__doc__',
'__file__',
'__loader__',
'__name__',
'__package__',
'__spec__',
'cclib',
'importChemLabQC',
'molOrbPlotter',
'np',
'pv',
'sys']
[8]:
print(out)
<module 'qc.wavefunction' from '/home/femtolab/python/chem/tools/chemlab/chemlab/qc/wavefunction.py'>
[9]:
# Test class
filename = r"/home/femtolab/ePS/XeF2/electronic_structure/xef2_SPKrATZP_rel_geom.log" # XeF2 Gamess file OK
mo = orbPlot.molOrbPlotter(chemPath = chemPath, fileIn = filename)
Import OK: Chemlab module <module 'qc.wavefunction' from '/home/femtolab/python/chem/tools/chemlab/chemlab/qc/wavefunction.py'>
Found electronic structure file: /home/femtolab/ePS/XeF2/electronic_structure/xef2_SPKrATZP_rel_geom.log
Read 3 atoms and 160 MOs
*** Grids set OK
StructuredGrid (0x7f2a027c6750)
N Cells: 117649
N Points: 125000
X Bounds: -2.937e+00, 2.937e+00
Y Bounds: -2.937e+00, 2.937e+00
Z Bounds: -2.937e+00, 2.937e+00
Dimensions: 50, 50, 50
N Arrays: 0
[10]:
mo
[10]:
<epsproc.vol.orbPlot.molOrbPlotter at 0x7f2a4097c510>
[11]:
mo.wavefunction
[11]:
<module 'qc.wavefunction' from '/home/femtolab/python/chem/tools/chemlab/chemlab/qc/wavefunction.py'>
Orb calcs¶
[12]:
mo.vol
[12]:
| StructuredGrid | Information |
|---|---|
| N Cells | 117649 |
| N Points | 125000 |
| X Bounds | -2.937e+00, 2.937e+00 |
| Y Bounds | -2.937e+00, 2.937e+00 |
| Z Bounds | -2.937e+00, 2.937e+00 |
| Dimensions | 50, 50, 50 |
| N Arrays | 0 |
[13]:
# Calculate orbital - default is HOMO
mo.calcOrb()
[14]:
mo.vol
[14]:
| Header | Data Arrays | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
[15]:
np.abs(mo.vol['36']).max()
[15]:
0.15967730310612654
[16]:
dir(mo.vol)
[16]:
['ALL_PIECES_EXTENT',
'AddObserver',
'AllocateCellGhostArray',
'AllocatePointGhostArray',
'AttributeTypes',
'BOUNDING_BOX',
'BlankCell',
'BlankPoint',
'BreakOnError',
'CELL',
'CELL_DATA_FIELD',
'CELL_DATA_VECTOR',
'CheckAttributes',
'ComputeBounds',
'CopyAttributes',
'CopyInformationFromPipeline',
'CopyInformationToPipeline',
'CopyStructure',
'Crop',
'DATA_EXTENT',
'DATA_EXTENT_TYPE',
'DATA_NUMBER_OF_GHOST_LEVELS',
'DATA_NUMBER_OF_PIECES',
'DATA_OBJECT',
'DATA_OBJECT_FIELD',
'DATA_PIECE_NUMBER',
'DATA_TIME_STEP',
'DATA_TYPE_NAME',
'DataHasBeenGenerated',
'DebugOff',
'DebugOn',
'DeepCopy',
'EDGE',
'EDGE_DATA_VECTOR',
'FIELD',
'FIELD_ACTIVE_ATTRIBUTE',
'FIELD_ARRAY_TYPE',
'FIELD_ASSOCIATION',
'FIELD_ASSOCIATION_CELLS',
'FIELD_ASSOCIATION_EDGES',
'FIELD_ASSOCIATION_NONE',
'FIELD_ASSOCIATION_POINTS',
'FIELD_ASSOCIATION_POINTS_THEN_CELLS',
'FIELD_ASSOCIATION_ROWS',
'FIELD_ASSOCIATION_VERTICES',
'FIELD_ATTRIBUTE_TYPE',
'FIELD_NAME',
'FIELD_NUMBER_OF_COMPONENTS',
'FIELD_NUMBER_OF_TUPLES',
'FIELD_OPERATION',
'FIELD_OPERATION_MODIFIED',
'FIELD_OPERATION_PRESERVED',
'FIELD_OPERATION_REINTERPOLATED',
'FIELD_OPERATION_REMOVED',
'FIELD_RANGE',
'FastDelete',
'FieldAssociations',
'FieldDataType',
'FieldOperations',
'FindAndGetCell',
'FindCell',
'FindPoint',
'GenerateGhostArray',
'GetActiveFieldInformation',
'GetActualMemorySize',
'GetAddressAsString',
'GetAssociationTypeAsString',
'GetAssociationTypeFromString',
'GetAttributeTypeForArray',
'GetAttributes',
'GetAttributesAsFieldData',
'GetBounds',
'GetCell',
'GetCellBounds',
'GetCellData',
'GetCellDims',
'GetCellGhostArray',
'GetCellNeighbors',
'GetCellPoints',
'GetCellType',
'GetCellTypes',
'GetCenter',
'GetClassName',
'GetCommand',
'GetData',
'GetDataDimension',
'GetDataObjectType',
'GetDataReleased',
'GetDebug',
'GetDimensions',
'GetExtent',
'GetExtentType',
'GetFieldData',
'GetGlobalReleaseDataFlag',
'GetGlobalWarningDisplay',
'GetInformation',
'GetLength',
'GetMTime',
'GetMaxCellSize',
'GetNamedFieldInformation',
'GetNumberOfCells',
'GetNumberOfElements',
'GetNumberOfPoints',
'GetPoint',
'GetPointCells',
'GetPointData',
'GetPointGhostArray',
'GetPoints',
'GetReferenceCount',
'GetScalarRange',
'GetUpdateTime',
'GlobalReleaseDataFlagOff',
'GlobalReleaseDataFlagOn',
'GlobalWarningDisplayOff',
'GlobalWarningDisplayOn',
'HasAnyBlankCells',
'HasAnyBlankPoints',
'HasAnyGhostCells',
'HasAnyGhostPoints',
'HasObserver',
'Initialize',
'InitializeObjectBase',
'InvokeEvent',
'IsA',
'IsCellVisible',
'IsPointVisible',
'IsTypeOf',
'Modified',
'NUMBER_OF_ASSOCIATIONS',
'NUMBER_OF_ATTRIBUTE_TYPES',
'NewCellIterator',
'NewInstance',
'ORIGIN',
'PIECE_EXTENT',
'POINT',
'POINT_DATA_FIELD',
'POINT_DATA_VECTOR',
'POINT_THEN_CELL',
'PrepareForNewData',
'ROW',
'Register',
'ReleaseData',
'RemoveAllObservers',
'RemoveNamedFieldInformation',
'RemoveObserver',
'RemoveObservers',
'SIL',
'SPACING',
'SafeDownCast',
'SetActiveAttribute',
'SetActiveAttributeInfo',
'SetDebug',
'SetDimensions',
'SetExtent',
'SetFieldData',
'SetGlobalReleaseDataFlag',
'SetGlobalWarningDisplay',
'SetInformation',
'SetPointDataActiveScalarInfo',
'SetPoints',
'SetReferenceCount',
'ShallowCopy',
'Squeeze',
'UnBlankCell',
'UnBlankPoint',
'UnRegister',
'UpdateCellGhostArrayCache',
'UpdatePointGhostArrayCache',
'VERTEX',
'VERTEX_DATA_VECTOR',
'__add__',
'__class__',
'__delattr__',
'__dict__',
'__dir__',
'__doc__',
'__eq__',
'__format__',
'__ge__',
'__getattribute__',
'__getitem__',
'__gt__',
'__hash__',
'__init__',
'__init_subclass__',
'__le__',
'__lt__',
'__module__',
'__ne__',
'__new__',
'__reduce__',
'__reduce_ex__',
'__repr__',
'__setattr__',
'__setitem__',
'__sizeof__',
'__str__',
'__subclasshook__',
'__this__',
'__vtkname__',
'__weakref__',
'_activate_texture',
'_active_scalars_info',
'_add_cell_array',
'_add_cell_scalar',
'_add_field_array',
'_add_field_scalar',
'_add_point_array',
'_add_point_scalar',
'_cell_array',
'_cell_arrays',
'_cell_bool_array_names',
'_clip_with_function',
'_field_array',
'_field_arrays',
'_field_bool_array_names',
'_from_arrays',
'_get_attrs',
'_ipython_key_completions_',
'_last_active_scalars_name',
'_load_file',
'_point_array',
'_point_arrays',
'_point_bool_array_names',
'_remove_array',
'_repr_html_',
'active_scalar',
'active_scalar_info',
'active_scalar_name',
'active_scalars',
'active_scalars_info',
'active_scalars_name',
'active_vectors',
'active_vectors_info',
'active_vectors_name',
'add_field_array',
'array_names',
'arrows',
'bounds',
'cast_to_unstructured_grid',
'cell_arrays',
'cell_centers',
'cell_data_to_point_data',
'center',
'center_of_mass',
'clear_arrays',
'clear_cell_arrays',
'clear_field_arrays',
'clear_point_arrays',
'clear_textures',
'clip',
'clip_box',
'clip_surface',
'compute_cell_quality',
'compute_cell_sizes',
'compute_gradient',
'connectivity',
'contour',
'copy',
'copy_meta_from',
'ctp',
'decimate_boundary',
'deep_copy',
'delaunay_3d',
'dimensions',
'elevation',
'extent',
'extract_cells',
'extract_edges',
'extract_geometry',
'extract_largest',
'extract_points',
'extract_selection_points',
'extract_surface',
'field_arrays',
'find_closest_point',
'get_array',
'get_data_range',
'glyph',
'head',
'interpolate',
'length',
'memory_address',
'merge',
'n_arrays',
'n_cells',
'n_points',
'n_scalars',
'number_of_cells',
'number_of_points',
'outline',
'outline_corners',
'overwrite',
'plot',
'plot_curvature',
'plot_over_line',
'point_arrays',
'point_data_to_cell_data',
'points',
'points_to_double',
'ptc',
'quality',
'references',
'rename_array',
'rename_scalar',
'rotate_x',
'rotate_y',
'rotate_z',
'sample',
'sample_over_line',
'save',
'scalar_names',
'select_enclosed_points',
'set_active_scalar',
'set_active_scalars',
'set_active_vectors',
'shallow_copy',
'slice',
'slice_along_axis',
'slice_along_line',
'slice_orthogonal',
'split_bodies',
'streamlines',
'surface_indices',
't_coords',
'texture_map_to_plane',
'textures',
'threshold',
'threshold_percent',
'transform',
'translate',
'triangulate',
'vectors',
'volume',
'warp_by_scalar',
'wireframe',
'x',
'y',
'z']
[17]:
mo.vol.array_names
[17]:
['36']
Orb plotting¶
[18]:
# With defaults
# Doesn't render plot unless called again?
mo.plotOrb()
[19]:
mo.pl.show()
# Inconsistent cmaps over geometries here? Way to rename and fix?
[20]:
# With pyVista plotter (non-interactive - better for web export?)
mo.plotOrb(interactive=False)
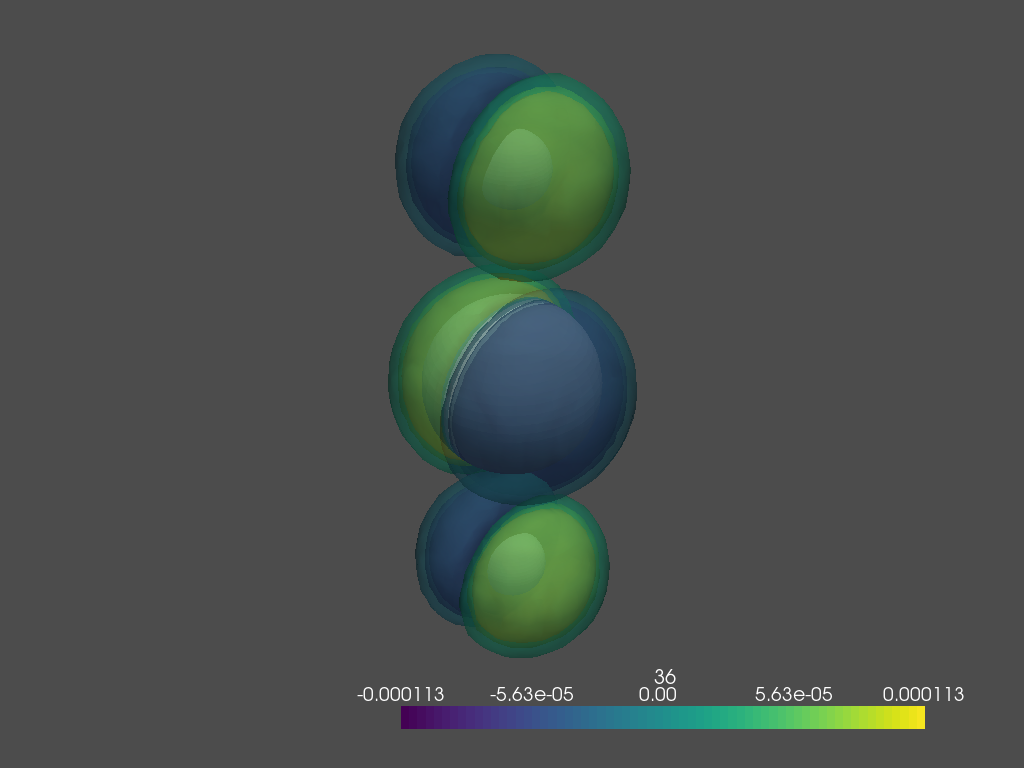
Coord scratch¶
[21]:
mo.data.atomcoords.max()
[21]:
1.9373215782
[22]:
mo.data.atomcoords
[22]:
array([[[ 0. , 0. , 0. ],
[ 0. , 0. , -1.929 ],
[ 0. , 0. , 1.929 ]],
[[-0. , 0. , -0. ],
[ 0. , -0. , -1.935778 ],
[ 0. , 0. , 1.935778 ]],
[[-0. , 0. , -0. ],
[ 0. , -0. , -1.93732158],
[ 0. , 0. , 1.93732158]]])
[23]:
mo.data.atomcoords[-1].min(axis = 0)
[23]:
array([-0. , 0. , -1.93732158])
[24]:
test = np.array([[1,2,3],[1,5,6], [-1,4,9]])
extent = np.array([test.min(axis=0), test.max(axis=0)])
[25]:
extent
[25]:
array([[-1, 2, 3],
[ 1, 5, 9]])
[26]:
extent.min()
[26]:
-1
[27]:
test2 = extent.copy()
test2[0,:] = test2.min()
test2[1,:] = test2.max()
[28]:
test2[0,:] -= 5
[29]:
test2
[29]:
array([[-6, -6, -6],
[ 9, 9, 9]])
[30]:
test3 = [-1, 5]
np.c_[np.ones(3)*test3[0], np.ones(3)*test3[1]].T
[30]:
array([[-1., -1., -1.],
[ 5., 5., 5.]])
[31]:
np.asarray(np.array(test3))
[31]:
array([-1, 5])
[32]:
axes = []
for col in extent.T:
print(col)
axes.append(np.linspace(col[0],col[1]))
[-1 1]
[2 5]
[3 9]
[33]:
axes = np.asarray(axes)
[34]:
np.meshgrid(axes[0], axes[1], axes[2])
[34]:
[array([[[-1. , -1. , -1. , ..., -1. ,
-1. , -1. ],
[-0.95918367, -0.95918367, -0.95918367, ..., -0.95918367,
-0.95918367, -0.95918367],
[-0.91836735, -0.91836735, -0.91836735, ..., -0.91836735,
-0.91836735, -0.91836735],
...,
[ 0.91836735, 0.91836735, 0.91836735, ..., 0.91836735,
0.91836735, 0.91836735],
[ 0.95918367, 0.95918367, 0.95918367, ..., 0.95918367,
0.95918367, 0.95918367],
[ 1. , 1. , 1. , ..., 1. ,
1. , 1. ]],
[[-1. , -1. , -1. , ..., -1. ,
-1. , -1. ],
[-0.95918367, -0.95918367, -0.95918367, ..., -0.95918367,
-0.95918367, -0.95918367],
[-0.91836735, -0.91836735, -0.91836735, ..., -0.91836735,
-0.91836735, -0.91836735],
...,
[ 0.91836735, 0.91836735, 0.91836735, ..., 0.91836735,
0.91836735, 0.91836735],
[ 0.95918367, 0.95918367, 0.95918367, ..., 0.95918367,
0.95918367, 0.95918367],
[ 1. , 1. , 1. , ..., 1. ,
1. , 1. ]],
[[-1. , -1. , -1. , ..., -1. ,
-1. , -1. ],
[-0.95918367, -0.95918367, -0.95918367, ..., -0.95918367,
-0.95918367, -0.95918367],
[-0.91836735, -0.91836735, -0.91836735, ..., -0.91836735,
-0.91836735, -0.91836735],
...,
[ 0.91836735, 0.91836735, 0.91836735, ..., 0.91836735,
0.91836735, 0.91836735],
[ 0.95918367, 0.95918367, 0.95918367, ..., 0.95918367,
0.95918367, 0.95918367],
[ 1. , 1. , 1. , ..., 1. ,
1. , 1. ]],
...,
[[-1. , -1. , -1. , ..., -1. ,
-1. , -1. ],
[-0.95918367, -0.95918367, -0.95918367, ..., -0.95918367,
-0.95918367, -0.95918367],
[-0.91836735, -0.91836735, -0.91836735, ..., -0.91836735,
-0.91836735, -0.91836735],
...,
[ 0.91836735, 0.91836735, 0.91836735, ..., 0.91836735,
0.91836735, 0.91836735],
[ 0.95918367, 0.95918367, 0.95918367, ..., 0.95918367,
0.95918367, 0.95918367],
[ 1. , 1. , 1. , ..., 1. ,
1. , 1. ]],
[[-1. , -1. , -1. , ..., -1. ,
-1. , -1. ],
[-0.95918367, -0.95918367, -0.95918367, ..., -0.95918367,
-0.95918367, -0.95918367],
[-0.91836735, -0.91836735, -0.91836735, ..., -0.91836735,
-0.91836735, -0.91836735],
...,
[ 0.91836735, 0.91836735, 0.91836735, ..., 0.91836735,
0.91836735, 0.91836735],
[ 0.95918367, 0.95918367, 0.95918367, ..., 0.95918367,
0.95918367, 0.95918367],
[ 1. , 1. , 1. , ..., 1. ,
1. , 1. ]],
[[-1. , -1. , -1. , ..., -1. ,
-1. , -1. ],
[-0.95918367, -0.95918367, -0.95918367, ..., -0.95918367,
-0.95918367, -0.95918367],
[-0.91836735, -0.91836735, -0.91836735, ..., -0.91836735,
-0.91836735, -0.91836735],
...,
[ 0.91836735, 0.91836735, 0.91836735, ..., 0.91836735,
0.91836735, 0.91836735],
[ 0.95918367, 0.95918367, 0.95918367, ..., 0.95918367,
0.95918367, 0.95918367],
[ 1. , 1. , 1. , ..., 1. ,
1. , 1. ]]]),
array([[[2. , 2. , 2. , ..., 2. ,
2. , 2. ],
[2. , 2. , 2. , ..., 2. ,
2. , 2. ],
[2. , 2. , 2. , ..., 2. ,
2. , 2. ],
...,
[2. , 2. , 2. , ..., 2. ,
2. , 2. ],
[2. , 2. , 2. , ..., 2. ,
2. , 2. ],
[2. , 2. , 2. , ..., 2. ,
2. , 2. ]],
[[2.06122449, 2.06122449, 2.06122449, ..., 2.06122449,
2.06122449, 2.06122449],
[2.06122449, 2.06122449, 2.06122449, ..., 2.06122449,
2.06122449, 2.06122449],
[2.06122449, 2.06122449, 2.06122449, ..., 2.06122449,
2.06122449, 2.06122449],
...,
[2.06122449, 2.06122449, 2.06122449, ..., 2.06122449,
2.06122449, 2.06122449],
[2.06122449, 2.06122449, 2.06122449, ..., 2.06122449,
2.06122449, 2.06122449],
[2.06122449, 2.06122449, 2.06122449, ..., 2.06122449,
2.06122449, 2.06122449]],
[[2.12244898, 2.12244898, 2.12244898, ..., 2.12244898,
2.12244898, 2.12244898],
[2.12244898, 2.12244898, 2.12244898, ..., 2.12244898,
2.12244898, 2.12244898],
[2.12244898, 2.12244898, 2.12244898, ..., 2.12244898,
2.12244898, 2.12244898],
...,
[2.12244898, 2.12244898, 2.12244898, ..., 2.12244898,
2.12244898, 2.12244898],
[2.12244898, 2.12244898, 2.12244898, ..., 2.12244898,
2.12244898, 2.12244898],
[2.12244898, 2.12244898, 2.12244898, ..., 2.12244898,
2.12244898, 2.12244898]],
...,
[[4.87755102, 4.87755102, 4.87755102, ..., 4.87755102,
4.87755102, 4.87755102],
[4.87755102, 4.87755102, 4.87755102, ..., 4.87755102,
4.87755102, 4.87755102],
[4.87755102, 4.87755102, 4.87755102, ..., 4.87755102,
4.87755102, 4.87755102],
...,
[4.87755102, 4.87755102, 4.87755102, ..., 4.87755102,
4.87755102, 4.87755102],
[4.87755102, 4.87755102, 4.87755102, ..., 4.87755102,
4.87755102, 4.87755102],
[4.87755102, 4.87755102, 4.87755102, ..., 4.87755102,
4.87755102, 4.87755102]],
[[4.93877551, 4.93877551, 4.93877551, ..., 4.93877551,
4.93877551, 4.93877551],
[4.93877551, 4.93877551, 4.93877551, ..., 4.93877551,
4.93877551, 4.93877551],
[4.93877551, 4.93877551, 4.93877551, ..., 4.93877551,
4.93877551, 4.93877551],
...,
[4.93877551, 4.93877551, 4.93877551, ..., 4.93877551,
4.93877551, 4.93877551],
[4.93877551, 4.93877551, 4.93877551, ..., 4.93877551,
4.93877551, 4.93877551],
[4.93877551, 4.93877551, 4.93877551, ..., 4.93877551,
4.93877551, 4.93877551]],
[[5. , 5. , 5. , ..., 5. ,
5. , 5. ],
[5. , 5. , 5. , ..., 5. ,
5. , 5. ],
[5. , 5. , 5. , ..., 5. ,
5. , 5. ],
...,
[5. , 5. , 5. , ..., 5. ,
5. , 5. ],
[5. , 5. , 5. , ..., 5. ,
5. , 5. ],
[5. , 5. , 5. , ..., 5. ,
5. , 5. ]]]),
array([[[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
...,
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ]],
[[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
...,
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ]],
[[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
...,
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ]],
...,
[[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
...,
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ]],
[[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
...,
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ]],
[[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
...,
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ],
[3. , 3.12244898, 3.24489796, ..., 8.75510204,
8.87755102, 9. ]]])]
[35]:
mo.data.homos[-1]
[35]:
35
[36]:
mo.data.mocoeffs[2].shape
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
<ipython-input-36-b4a687dcf1db> in <module>
----> 1 mo.data.mocoeffs[2].shape
IndexError: list index out of range
Isovals scratch¶
[ ]:
isoValsPC = np.array([0.1, 5, 10])
isoLevels = 6
item = '36'
limitVals = [mo.vol[item].min(), mo.vol[item].max(), np.abs(mo.vol[item]).mean()]
# np.r_[isoValsPC* limitVals[2], isoValsPC* (-1*limitVals[2])] # This is OK with lists until *-1
np.r_[isoValsPC, -isoValsPC]*limitVals[2] # This is
[ ]:
np.asarray(None)
[ ]:
isoValsOrb = np.linspace(-limitVals[2], limitVals[2], isoLevels)
[ ]:
len(isoValsOrb)
[ ]:
mo.vol.contour(isoValsOrb, scalars = item)
[ ]:
isoValsPC = None
if isoValsPC is None:
isoValsOrb = np.r_[isoValsPC, -isoValsPC] * limitVals[2] # Set PC vals from mean?
print(isoValsOrb)
[ ]:
for item in mo.vol.array_names:
print(item)
[ ]:
isoValsAbs = np.array(None)
[ ]:
isoValsAbs
[ ]: